Scientists make important discovery around antibiotic resistance
Researchers have discovered why superbugs are resistant to certain antibiotics.
By Lana HaightDifferent types of antibiotics.
Researchers from McGill University have discovered why superbugs are resistant to certain antibiotics and are one step closer to finding new treatments.

“Superbugs have different tricks up their sleeves on how to resist antibiotics. We became interested in looking at how they resist these macrolide antibiotics, like erythromycin ,” said Dr. Albert Berghuis, chair of the Department of Biochemistry at McGill University’s Faculty of Medicine.
Macrolide antibiotics are the fourth largest class of antibiotics and are prescribed for a variety of infectious diseases, including upper respiratory tract, skin and soft tissue infections. Erythromycin and other macrolide antibiotics are also prescribed as an alternative to penicillin. In 2009, sales of macrolide antibiotics in the U.S. topped $4.8 billion.
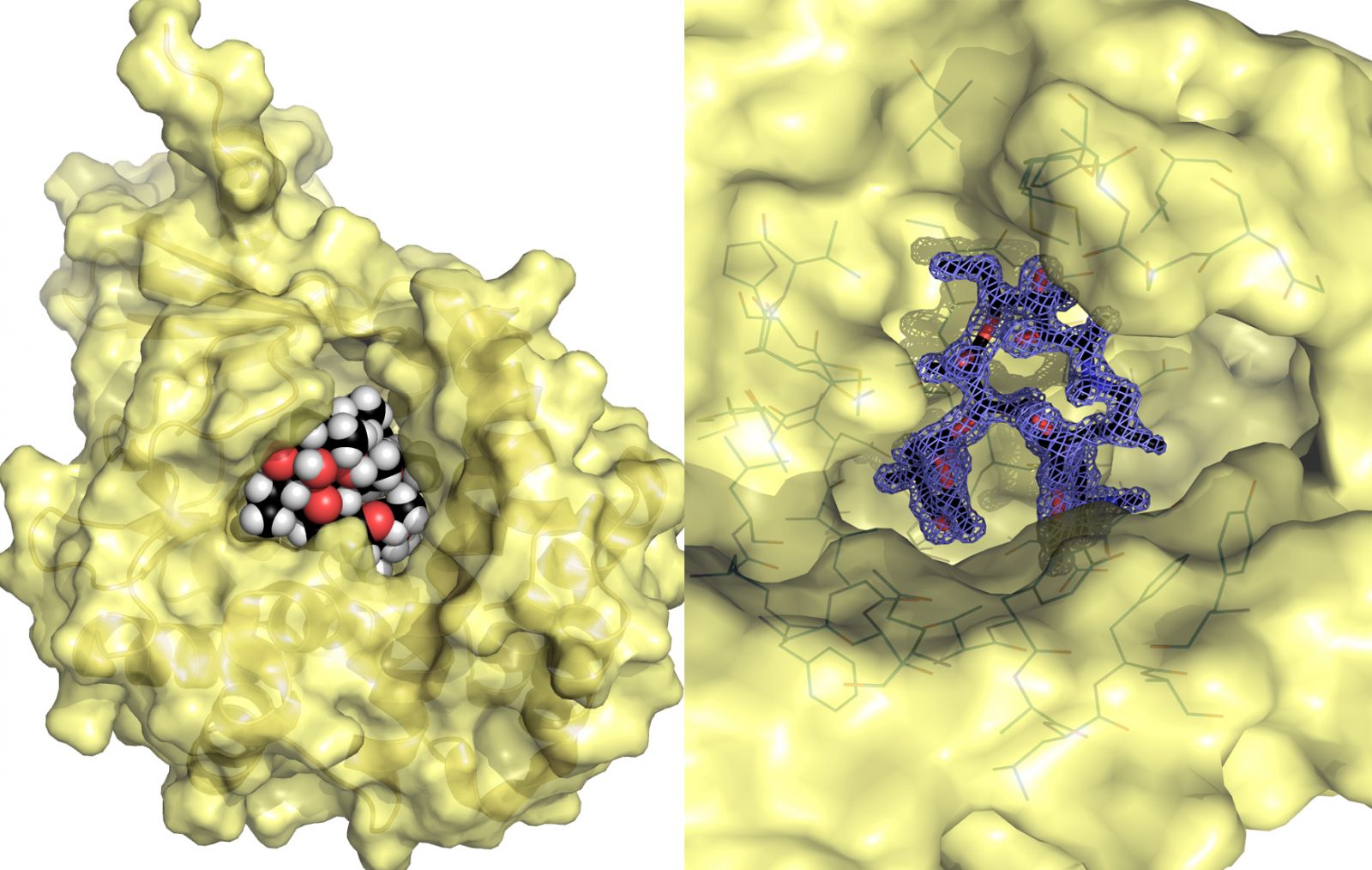
Dr. Berghuis and his colleagues conducted research using the intensely bright light at the Canadian Light Source, the only synchrotron in Canada, and were able to analyze at the atomic level enzymes called kinases. The research group’s findings have just been published in the journal Structure.

It’s the enzymes in the bacteria that have the ability to chemically modify antibiotics. The enzymes take a molecule that has a phosphate group in it and they stick that phosphate group on to something else, explained Dr. Berghuis.
“In the kinases that we studied, they stick the phosphate group on to the macrolide antibiotic and that little change is just enough to make them no longer an antibiotic. When that happens, it becomes a useless compound and the bacteria lives happily ever after which is not good for us.”
Dr. Berghuis compared the process of an enzyme attacking an antibiotic to using a coloured marker. When the lid is not on the marker, the marker can be used to colour on paper covering whatever is on the paper, just as a macrolide antibiotic covers a bacteria and kills it. You put a lid on that marker and it can no longer be used to colour. Similarly, when the bacteria’s enzyme attaches a chemical to the antibiotic, it also is rendered useless.
The images and data collected at the Canadian Light Source provided the essential information in out-tricking superbugs.
“We now know in exquisite detail how these macrolides interact with the enzymes. That exquisite detail is critical if you want to make a version of these macrolides that don’t bind anymore to the enzymes. Without that information, this whole plan goes out the window. That’s what the CLS allowed us to do.”
Now that researchers know how this process works, the next step will take them back to the chemistry laboratory.
“With this knowledge, we can design a slightly different antibiotic that the enzymes can no longer recognize. We can make a variant of the macrolide that is resistant to the resistance.”
Dr. Berghuis estimates that the development of new macrolide antibiotics will take two to three years and then more time will be needed for clinical trials before they are readily available. In the meantime, he says everyone needs to do their part in the battle against superbugs by using antibiotics appropriately. Overuse and improper use, not completing medication, have contributed to development of superbugs and their antibiotic resistance.
Fong, Desiree H., David L. Burk, Jonathan Blanchet, Amy Y. Yan, and Albert M. Berghuis. "Structural Basis for Kinase-Mediated Macrolide Antibiotic Resistance." Structure (2017). DOI: 10.1016/j.str.2017.03.007
To arrange an interview, contact:
Victoria Schramm
Communications Coordinator
Canadian Light Source
306-657-3516
victoria.schramm@lightsource.ca
