Scientists break record while battling antibiotic resistance
Researchers from McGill University used the CLS to determine the physical structure of largest complex ever measured using the CMCF beamline.
By Rowan HollingerDr. Albert Berghuis
Drug-resistant diseases could cause up to 10 million deaths a year by 2050, according to the World Health Organization. Scientists used the Canadian Light Source (CLS) at the University of Saskatchewan to better understand how current antibiotics work and how we might curb bacterial resistance to these life-saving drugs.
Many new antibiotics are able to kill infection-causing bacteria by binding to these bacteria’s ribosomes, which are the essential machines that make proteins. In order to see exactly what antibiotics do at an atomic level, researchers from McGill University used the CLS to determine the physical structure of a ribosome as it interacted with one of the newest antibiotics.
To understand how some bacteria are already resistant to this new antibiotic, they also determined how the drug interacts with a key bacterial enzyme that causes the resistance. The results were recently published in Nature Communications Biology.
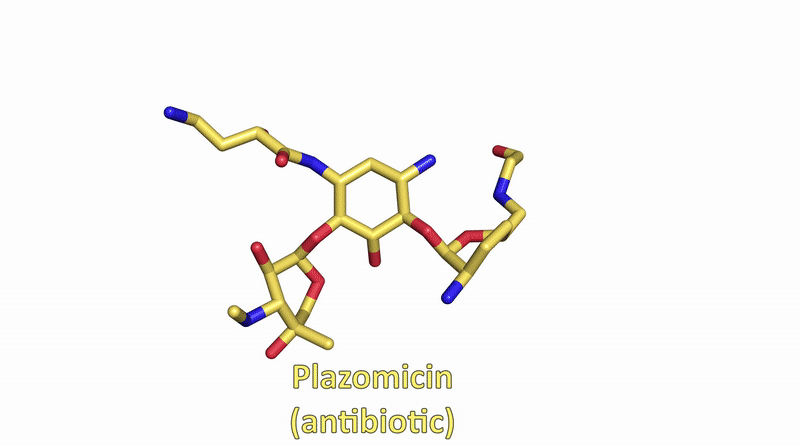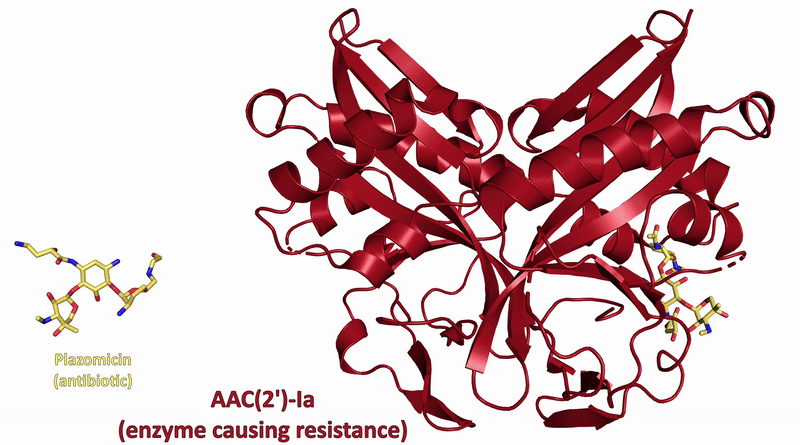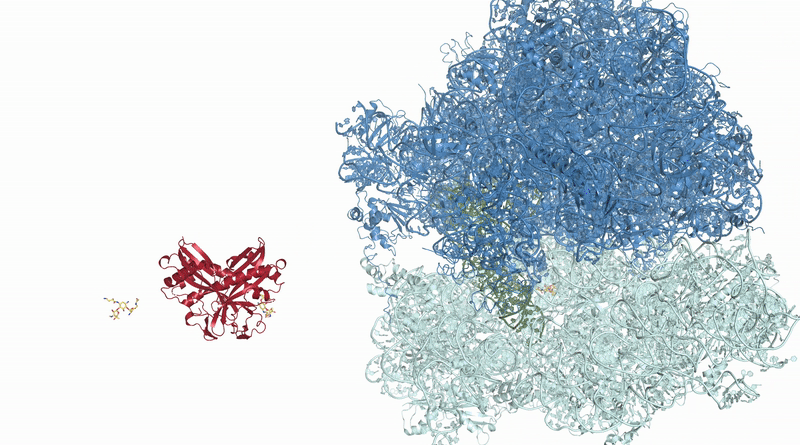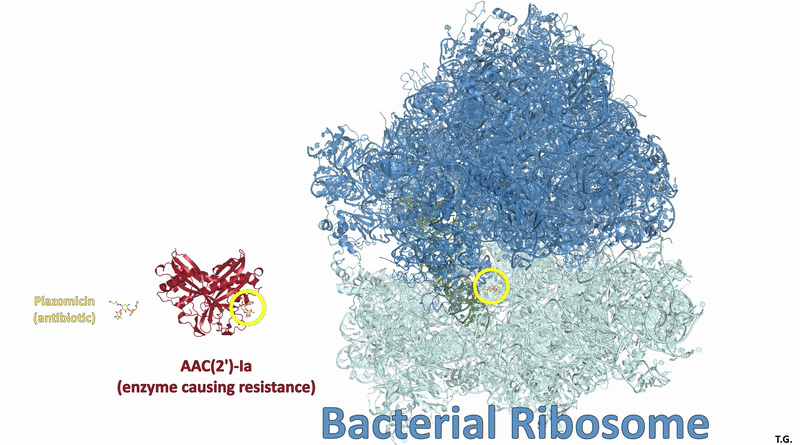
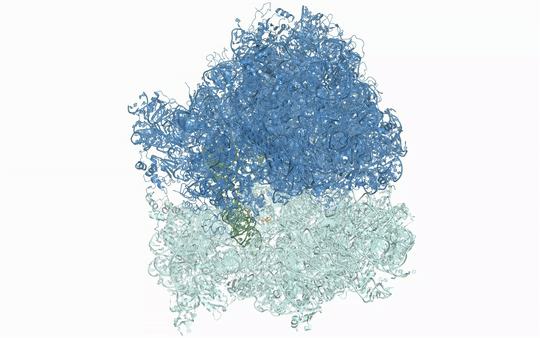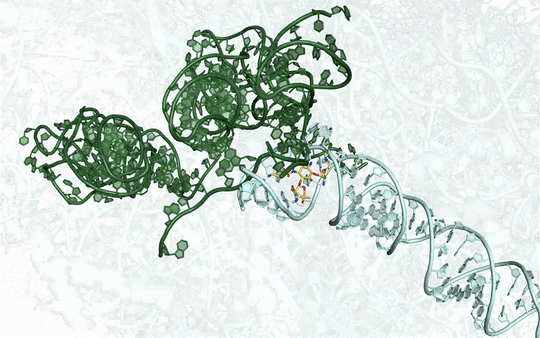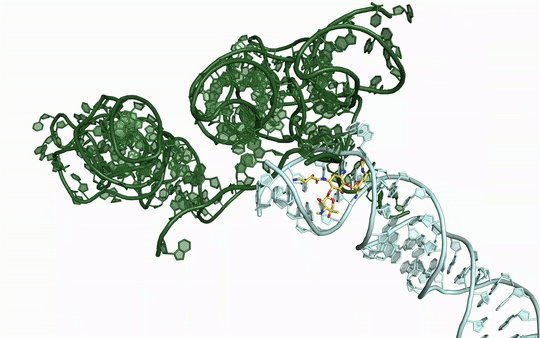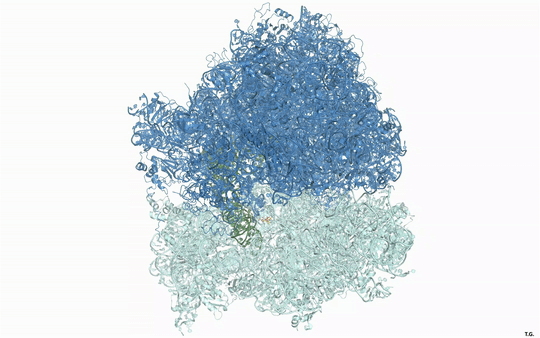
Visualizing the antibiotic bound to the ribosome, which is a complex with 300,000 atoms, was a feat that took the team roughly five years to complete. In the process, the scientists broke the record for the largest structure ever analyzed using the CMCF beamline at the CLS, which is the only facility of its kind in Canada. The previous record, set in 2013, was for a structure six times smaller.
“The antibiotic we studied is made of about 40 atoms – just a little bigger than one sugar molecule,” explained lead researcher Dr. Albert Berghuis, Chair of the Department of Biochemistry at McGill University’s Faculty of Medicine and Health Sciences. “It’s a bit crazy that we had to figure out 300,000 atoms in order to know what those 40 atoms in the antibiotic are doing.”
Berghuis described the research process as both fun and very challenging.
“The people at the CMCF beamline deserve a lot of credit,” said Berghuis. “They tried probably 50 to 80 samples, and only a few of them worked really well.” He also credited the hard work co-authors Tolou Golkar and Angelia Bassenden from McGill put into the research.
By determining the structural basis for antibiotic action and resistance, their work will make it easier to create and test antibiotics in the future.
This research builds on work published in 2017 and 2021 that the team conducted in collaboration with the CLS. With that knowledge, they made sure to analyze the structure of an antibiotic as it interacted with a ribosome as well as this enzyme. According to Berghuis, figuring out both parts will be critical to making better antibiotics.
“We will now want to make a next-generation drug that still binds to the ribosome, but no longer to enzymes that will cause resistance,” said Dr. Berghuis. “That’s why we needed both pieces of information. Now we have that picture, we can make changes.”
Their findings will allow researchers to test the interaction between ribosomes and next-generation antibiotics in great detail and in a much shorter time frame.
“We have figured out all the problems. We have jumped over all the hurdles,” said Berghuis, who estimates that replicating the research with a new form of antibiotic would not take another five years but roughly a tenth of that time.
The team hopes that these discoveries can fast-track the development of more effective antibiotics.
Golkar, Tolou, Angelia V. Bassenden, Krishnagopal Maiti, Dev P. Arya, T. Martin Schmeing, and Albert M. Berghuis. "Structural basis for plazomicin antibiotic action and resistance." Communications biology 4, no. 1 (2021): 1-8. https://doi.org/10.1038/s42003-021-02261-4
To arrange an interview, contact:
Victoria Schramm
Communications Coordinator
Canadian Light Source
306-657-3516
victoria.schramm@lightsource.ca
